| 送交者: 苦难与荣耀[☆★★声望品衔11★★☆] 于 2021-07-05 3:43 已读 33351 次 | 苦难与荣耀的个人频道 |
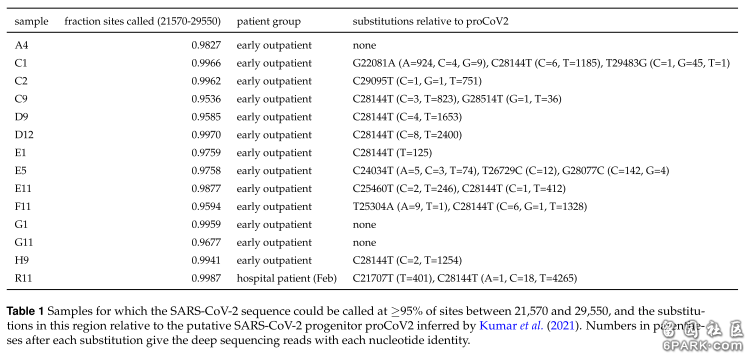
长标题:美国病毒学家Bloom新冠溯源研究恢复、依据的基础数据,是武汉大学“纳米孔靶向测序检测”技术研发、测试过程中测序、输出的新冠病毒基因序列片断,这些基因序列片断准确度欠佳,存在大量的核苷酸测序缺失。 6park.comLong title: the basic data on which American virologist Bloom's origination research of Sars-Cov-2 based, is the Sars-Cov-2 sequence fragments which sequenced and output by the "nanopore targeted sequencing detection" technology of Wuhan University, these sequence fragments is produced during the developing and testing process, their accuracy is not good enough for gene and Sars-Cov-2’s origination research。There are lots of nucleotide miss in the Sars-Cov-2 sequence fragments Bloom recovered. 6park.com本文是 "病毒学家Bloom溯源论文的基础数据问题"的一个中英对照版本,写作中借助了谷歌翻译。原中文版地址为: https://club.6parkbbs.com/bolun/index.php?app=forum&act=threadview&tid=15952734 6park.comThis article is the Chinese-English version of the article "病毒学家Bloom溯源论文的基础数据问题",Google Translate was used in my writing. The address of original Chinese version is as follow: 6park.com对美国病毒学家Jesse Bloom的新冠溯源论文作了初步研究,发现了一些疑点、缺陷和错误。本文整理的是与论文的基础研究数据相关的一组问题。成文仓促,请各位网友批评、指正。 6park.comPreliminary research has been done on the Sars-Cov-2 origination paper of American virologist Jesse Bloom, and some doubts, defects and errors have been found. This article sorts out a set of questions related to the basic research data of the thesis. Written hastily, looking forward to your criticisms and corrections。 6park.com2021年6月22日,美国西雅图贺勤森癌症研究中心(Fred Hutchinson Cancer Research Center,福瑞德·哈金森癌症研究中心)的病毒学家杰西·布鲁姆(Jesse Bloom)在生物学预印本平台biorxiv发布了如下论文:Recovery of deleted deep sequencing data sheds more light on the early Wuhan SARS-CoV-2 epidemic(恢复删除的深度测序数据为新冠病毒在武汉的早期流行提供了更多信息) https://www.biorxiv.org/content/10.1101/2021.06.18.449051v1 https://www.biorxiv.org/content/10.1101/2021.06.18.449051v1.full 6park.comOn June 22, 2021, virologist Jesse Bloom released the following paper on the preprint platform biorxiv : Recovery of deleted deep sequencing data sheds more light on the early Wuhan SARS-CoV-2 epidemic 6park.comJesse Bloom is a virologist of the Fred Hutchinson Cancer Research Center in Seattle, USA. 6park.com本文将说明以下6点: 6park.comThis article will explain the following 6 points: 6park.com1、Bloom恢复的基因序列,是武汉大学“纳米孔靶向测序检测”技术研发过程中产生的测试数据; 6park.com1. The Sars-Cov-2 sequences restored by Bloom is the testing data generated during the research and development of the "Nanopore Targeted Sequencing Detection" technology of Wuhan University; 6park.com2、关于数据是如何删除的,还存在疑点,还不能下定论。一种可能是:武汉大学在完成相关项目后,向NIH请求撤回(删除或移走)了他们在研发、测试阶段输出的,提交到SRA保存的新冠病毒基因序列片断; 6park.com2. Regarding how the data was deleted, there are still doubts and no conclusion can be drawn. One possibility is that after completing the relevant project, Wuhan University requested NIH to withdraw the Sars-Cov-2 sequence fragments that they exported during the R&D and testing stages and submitted to the SRA for preservation; 6park.com3、“纳米孔靶向测序检测”技术是qPCR病毒检测技术的替代技术,其核心功能是病毒检测,而非病毒测序。检测样本中是否存在目标病毒是这一技术的首要功能,对病毒进行部分测序并输出测序结果是其辅助功能。而且,这一技术测序、输出的不是新冠病毒的全基因组序列,只是与检测功能相关的全基因组序列的若干片断; 6park.com3. "Nanopore targeted sequencing detection" technology is an alternative to qPCR virus detection technology, and its core function is virus detection, not virus sequencing. Detecting the presence of the target virus in a sample is the primary function of this technology, and partially sequencing the virus and outputting the sequencing result is its auxiliary function. Moreover, the sequencing and output of this technology is not the whole genome sequence of Sars-Cov-2, but only several fragments of the whole genome sequence related to the detection function; 6park.com4、“纳米孔靶向测序检测”技术输出的基因序列,允许更大误差和更高错误率,不应将它们等同于专门测序得到的,可向国际生物数据库正式提交的,可用于严谨基因科学研究的,权威性的基因序列。即使这些基因序列片断仍存在科学研究价值,在将它们用于对准确性要求极高的基因科学研究,特别是新冠溯源研究时,也应当非常审慎,也应当对其中的测序错误可能产生的影响、误导进行充分的分析、评估。 6park.com4. The gene sequences output by the "Nanopore Targeted Sequencing Detection" technology allow larger errors and higher error rates. They should not be equated with the authoritative gene sequence obtained by special sequencing,the latter can be formally submitted to the international biological database,taken as base data of rigorous gene research. Even if these sequence fragments still have scientific research value, they should be used very cautiously when they are used in genetic scientific research that requires extremely high accuracy, especially the origination research of Sars-Cov-2, careful and adequate analysis and evaluation should be made to avoid being misled by the sequencing errors. 6park.com5、Bloom恢复的序列片断证明,武汉大学“纳米孔靶向测序检测”技术的测序结果中存在大量的核苷酸测序缺失。 6park.com5. The sequence fragments recovered by Bloom prove that there are a large number of nucleotide sequencing deletions in the sequencing results of the "Nanopore Targeted Sequencing Detection" technology of Wuhan University. 6park.com6、武汉大学“纳米孔靶向测序检测”技术输出的欠精确、可靠的研发、测试数据,被Bloom拿来充当了其溯源论文的基础数据。是否采取措施规避了基础数据中的测序错误?测序错误可能对研究过程及论文结论造成什么样的影响或误导?Bloom在论文中没有进行讨论、评估。 6park.com6. The inaccurate and reliable R&D and test data output by Wuhan University's "Nanopore Targeted Sequencing Detection" technology was used by Bloom as the basic data in its paper about origination of Sars-Cov-2. Have measures been taken to avoid the effects of sequencing errors in the basic data? What kind of influence or misleading might the sequencing error have on the research process and the conclusion of the paper? Bloom did not discuss or evaluate in the paper. 6park.com下面对上述6点予以展开。 6park.comThe above 6 points will be expanded below. 6park.comBloom在论文中说,他通过谷歌云恢复了去年6月被删除的一些测序数据,他通过这些数据重建了13个新冠病毒的基因序列。论文提供了这13个基因序列的下载地址: https://github.com/jbloom/SARS-CoV-2_PRJNA612766/raw/main/results/consensus/consensus_seqs.csv 6park.comBloom said in the paper, he recovered some sequencing data deleted in June last year, and he reconstructed 13 Sars-Cov-2 sequences through the data. The paper provides the download address of these 13 sequences: 6park.com需要指出的是,Bloom重建的13个序列都不是完整的新冠病毒全基因组序列,每个序列都是新冠全基因组序列一大一小的两个片断。Bloom未得到全基因组序列,不是因为他恢复得不完全,而是因为他恢复的数据本来就是基因组片断。这些新冠基因组片断来自武汉大学“纳米孔靶向测序检测”(Nanopore Targeted Sequencing,简称NTS)项目,是这一项目研发、测试过程中产生、输出的。NTS技术不需要检测新冠病毒的全基因组,只需要比对、测序全基因组中的约12个区段。 6park.comIt should be noted that 13 sequences reconstructed by Bloom are not complete Sars-Cov-2 genome sequences, each sequence contains two fragments, one long,another very short. Bloom did not get the whole genome sequences, not because he recovered incomplete, but because the data he recovered were originally genome pieces. These Sars-Cov-2 genomic pieces come from Wuhan University Nanopore Targeting Sequencing(referred to as NTS) project, produced and output during the developing、testing process of this project. NTS technology does not need to detect the whole genome of Sars-Cov-2, only about 12 sections need to be compared and sequencing. 6park.com后续内容将涉及一个生物信息数据库SRA。SRA,即Sequence Read Archive(序列读取档案)是NIH(National Institutes of Health,美国国立卫生研究院)管理、维护的两个生物学数据库之一,另一个是GenBank。 6park.comSubsequent content will involve a biological information database SRA. SRA, the Sequence Read Archive is one of the two biological databases of NIH (National Institutes Of Health), the other is GenBank. 6park.comNTS项目使用SRA为研发的数据存储平台,它在SRA上的项目代号为Bio-Project PRJNA612766。Bloom论文中说,截止2020年3月30日,PRJNA612766项目共向SRA提交了282 份数据,其中241份数据的相关信息被一位名叫Carlos Farkas的科学家(等人)整理在一个Excel表格中,Bloom就是依据表格中的信息恢复了部分数据并重建了13个序列片断。Bloom恢复的数据只是PRJNA612766即武汉大学NTS项目提交数据的很小一部分。我粗略地浏览了该表格,NTS项目最早的数据提交时间应该是2020年1月15日。顺便附上Bloom论文提供的Farkas表格的下载地址。 https://web.archive.org/web/20210502130356/https://dfzljdn9uc3pi.cloudfront.net/2020/9255/1/Supplementary_Table_1.xlsx. 6park.comThe NTS project uses SRA as their data storage platform, its SRA project code is Bio-Project PRJNA612766.In the bloom paper, as of March 30, 2020, PRJNA612766 projects submitted 282 data to SRA, 241 of them were organized by scientists called Carlos Farkas (et al.) In an Excel form. In accordance with the information in the table, Bloom recovered and reconstructed 13 sequence pieces. The data Bloom recovered are only a small part of PRJNA612766,the NTS project of Wuhan University. I roughly browsed the form, the earliest data submission time of the NTS project should be January 15, 2020. By the way, the download address of the FARKAS table provided by the bloom paper. 6park.comBloom重建的13个序列片断的对象时间戳都是2020年2月15日。Bloom推测,对象时间戳可能是指数据上传到SRA的时间。也就是说,Bloom恢复的数据应该是同一批数据,它们都是2020年2月15日这天上传到SRA的。 6park.comThe target timestamps of the 13 sequence fragments Bloom reconstructed are all February 15, 2020. Bloom speculates that an object timestamp may refer to the time of data submitted to SRA. That is, the data recovered by Bloom should be the same batch of data, which are all uploaded to SRA on February 15, 2020. 6park.comBloom自已通过谷歌云恢复、重建了13个序列片断。其实,他可以通过NIH,利用SRA的数据库备份系统恢复所有相关数据。论文没有提及这一数据恢复途径,没有解释Bloom为什么不通过这一途径完整地、更可靠地恢复数据。 6park.comBloom restored 13 sequence fragments through Google Cloud by himself. In fact, he can restore all relevant data by NIH, using SRA's database backup system. The papers did not mention this data recovery approach, and did not explain why Bloom did not restore data completely and more reliably by this way. 6park.comBloom论文引用的武汉大学NTS(纳米孔靶向测序检测)研究的相关论文为:Nanopore target sequencing for accurate and comprehensive detection of SARS-CoV-2 and other respiratory viruses(纳米孔靶向测序可准确、全面检测 SARS-CoV-2 和其他呼吸道病毒) https://www.medrxiv.org/content/10.1101/2020.03.04.20029538v1.full-text 6park.comThe related papers of NTS (nanopore target sequencing detection) studies quoted by bloom papers is as:Nanopore target sequencing for accurate and comprehensive detection of SARS-CoV-2 and other respiratory viruses 6park.com这一论文投稿到预印版平台medRxiv的时间是2020年2月29日,正式发布时间是2020年3月6日。 6park.comThe time of this paper submitted to the pre-printed platform Medrxiv was February 29, 2020, and it officially released on March 6, 2020. 6park.com2020年3月4日,中国人民日报和新华网报道了这一新技术: 武汉大学研发纳米孔靶向测序检测方法 http://www.xinhuanet.com/science/2020-03/04/c_138841605.htm 6park.comOn March 4, 2020, the People's Daily and Xinhuanet reported this new technology: Wuhan University R & D nanopore target sequencing detection method 6park.com概要地介绍一下武汉大学“纳米孔靶向测序检测”技术(NTS)的功能特点,这将有助于我们作出进一步的正确判断: 6park.comThe below is the summary introduction to the functional characteristics of Wuhan University "Nanohole Targeting Detection" Technology (NTS), which will help us make further correct judgment: 6park.com1、相比传统的qPCR(quantitative polymerase chain reaction,实时荧光定量PCR,或实时定量聚合酶链反应)检测30%~50%的阳性检出率,NTS将阳性检出率提升了43.8%,达到约75%~94%; 6park.com1, compared to positive detection rate of 30% to 50% of the traditional QPCR (Quantitative Polymerase Chain Reaction,or real-time fluorescent PCR, or real-time quantitative polymerase chain), NTS increased the positive detection rate by 43.8% and achieve about 75% to 94%; 6park.com2、对于高浓度病毒样本,NTS仅需测序10分钟即可检测阳性,即使极低浓度病毒样本,也仅需测序4小时完成检测,从收到样本到出具结果,全程控制在6-10小时; 6park.com2. For high concentration virus samples, NTS only needs to sequencing for 10 minutes to detect positive, even if the virus sample is very low concentration, only 4 hours is needed to complete the detection, from receiving the sample to returning the results, only 6-10 hours is need for the full steps; 6park.com3、NTS可在测序后4小时内高敏感性、高准确性地同时检测SARS-CoV-2和其他10大类、40余种呼吸道病毒; 6park.com3, NTS can detect SARS-COV-2 and other 10 categories, more than 40 kinds of respiratory viruses with high sensitivity and high accuracy within 4 hours after sequencing. 6park.com4、NTS最低检测敏感度是广泛使用的传统qPCR的100倍; 6park.com4, The minimum detection sensitivity of NTS is 100 times that of traditional QPCR extensively used; 6park.com5、NTS还可输出检测样本中病原体(如新冠病毒)的基因组序列片断,可用于考察新冠病毒基因组的变异情况,监控病毒变异引起的毒性与传播能力改变。这是qPCR所没有的功能,qPCR病毒检测只作基因比对,不作基因测序,不记录、输出任何基因组序列。 6park.com5. NTS also outputs gene sequence fragments for the detecting pathogens (such as Sars-Cov-2) in the sample, which can be used to investigate the mutation of Sars-Cov-2, and monitor the variation of the toxicity and communication capacity caused by viral mutation. This is the function that QPCR does not have.QPCR only performs gene comparison between sample virus and target virus, does not make gene sequencing, does not record, and output any genomic sequence. 6park.com6、NTS所需的纳米孔测序平台对实验室要求不高,其中最小测序仪MinION是便携式的,因此NTS也适合不同级别的医院使用。 6park.com6. The Nanohole-sequencing platform has no very high requirement on laboratory conditions,specially, its minimum sequencer named as MinION is portable, so NTS is also suitable for different levels of hospitals. 6park.com由上述功能细节可知,NTS技术是面向、服务于医院和普通实验室的;NTS技术对实验室条件要求不高,这应该暗示着它并不能胜任有着极高精度的专业测序;第5组功能是锦上添花性质的,该组部分功能很可能有夸大其词的噱头成份。一个仓促研发的产品,如果号称拥有众多强大功能,那很可能意味着,至少,它的相当一部分功能是不尽善尽美的。 6park.comAccording to the above functional details, NTS technology is facing, serving hospitals and ordinary laboratories; NTS technology has no high requirement on laboratory conditions, which should imply that it does not compete with extremely high precision professional sequencing; The fifth group functions is very likely to be exaggerated. A rush-developed product, If it claims to have a lot of powerful features, it likely means that, at least,its considerable parts of the functionality is not perfect. 6park.com下面讨论数据撤回、删除的有关情况。 6park.comThe following discussion is about the relevant situation of data withdrawal and deletion. 6park.comBloom在论文中说,当他按照Carlos Farkas论文和表格的指引去查阅武汉大学提交到SRA的数据时,在 NCBI-SRA系统中已经搜索不到相关项目PRJNA612766了,用Farkas提供的Accession ID直接搜索该项目下的相关序列数据,搜索结果提示:序列数据已被删除。 6park.comBloom said in the paper that, when he check the data submitted by Wuhan University following the guidance of Carlos Farkas paper and form, the relevant project PRJNA612766 could not be searched in the NCBI-SRA system, and when he search this project relevant data directly by the Accession ID provided by Farkas, the search result prompts: the sequence data has been deleted. 6park.comBloom说,SRA 被设计为深度测序数据的永久存档,上传数据到SRA后,只能通过向 SRA 工作人员发送电子邮件来删除数据。为说明可以通过发送邮件来请求删除数据,Bloom举了一个例子,他提供了一个Xiao姓科学家向SRA工作人员发送email,请求删除数据的电子邮件截图。 6park.comBloom said that SRA is designed as a permanent archive of deep sequencing data. After uploading the data to the SRA, the data can only be deleted by sending an email to the SRA staff. To illustrate that one can request deletion of data by sending an email, Bloom gave an example. He provided a screenshot of an email, the email is from a scientist surnamed Xiao, who sent this email to an SRA staff member requesting deletion of data. 6park.com6park.comBloom说,这位xiao姓科学家是一篇穿山甲冠状病毒论文的lead author。Bloom论文的“Literature Cited”(引用的文献)中指示了xiao姓科学家的论文: https://www.nature.com/articles/s41586-020-2313-x 6park.comBloom said that the scientist Xiao is the lead author of a pangolin coronavirus paper. The "Literature Cited" of the Bloom paper indicated the papers of the scientist Xiao: 6park.com由(穿山甲)论文作者列表可知,这个xiao姓的lead author或者是论文的通讯作者之一,华南农业大学特聘教授肖立华,或者是论文的第一作者Kangpeng Xiao。 6park.comAccording to the authors list of the pangolin related paper, the lead author with the surname Xiao is either one of the corresponding authors of the paper, Lihua Xiao, a distinguished professor of South China Agricultural University, or Kangpeng Xiao, the first author of the paper. 6park.comBloom提供Xiao姓科学家的email截图是为了例证:SRA数据可通过email请求撤回、删除。由截图可见,Xiao姓科学家删除的项目与武汉大学的NTS项目二者的BioProject ID不同,它们是两个不同的项目。 6park.comThe purpose that Bloom provided the email screenshot of the scientist Xiao is to illustrate: SRA data can be withdrawn or deleted via email request. It can be seen from the screenshot that the BioProject ID of the project deleted by the scientist Xiao is different from the NTS project of Wuhan University. They are two different projects. 6park.comBloom是怎么得到Xiao姓科学家邮件截图的?Bloom也未作说明。邮件截图是NIH提供的吗?如果是的话,NIH为什么不直接提供武汉大学PRJNA612766项目(即“纳米孔靶向测序检测”项目)相关的邮件,而要提供一个无关项目的邮件呢?如果邮件不是NIH提供的,那么,如此私密的信息,Bloom又是怎么得到的呢?Bloom和xiao姓科学家有什么直接或间接的关系吗? 6park.comHow did Bloom get the email screenshot of scientist Xiao? Bloom also didn't say anything. Are the email screenshots provided by NIH? If so, why doesn't NIH directly provide email related to Wuhan University's PRJNA612766 project, but give email of unrelated project? If the email was not provided by NIH, how did Bloom get such private information? Is there any direct or indirect relationship between Bloom and the scientist Xiao? 6park.com这些疑点暂时无法得到澄清。至少有以下两种可能性: 1、xiao姓科学家的邮件是NIH提供的,但NIH未提供或未能提供武汉大学PRJNA612766项目组人员请求删除数据的邮件; 2、xiao姓科学家的邮件是Bloom通过私人渠道获得的。在2018回国工作,被华南农业大学特聘前,肖立华在美国学习、工作了至少18年,在美国CDC工作了14年。 6park.comThese doubts cannot be clarified temporarily. There are at least two possibilities: 1. The email of the scientist Xiao was provided by NIH, but NIH did not provide or failed to provide the email by which the team member of PRJNA612766 project of Wuhan University requesting to delete the data; 2. The email of the scientist surnamed Xiao was obtained by Bloom through private channels. Before returning to work in China in 2018, and being specially hired by South China Agricultural University, Lihua Xiao studied and worked in the United States for at least 18 years, and worked for CDC of the United States for 14 years. 6park.com小结一下上述几段内容。Bloom称数据被删除了,称SRA数据只能通过发送email请求删除,他还举了一个其它项目通过这一途径请求删除数据的例子,但他未提供武汉大学项目组人员要求删除数据的直接证据。Bloom论文提供的信息,让我无法肯定,相关数据确实是武汉大学项目组人员要求删除的,我不能排除数据删除的其它可能性。 6park.comSummarize the above paragraphs. Bloom claimed that the data was deleted, saying that SRA data can only be deleted by sending an email request. He also cited an example of another project requesting data deletion through this approach, but he did not provide direct evidence that the Wuhan University project team requested the deletion of the data. The information provided in the Bloom paper makes me not sure that the relevant data was indeed deleted by the project team of Wuhan University, and I cannot rule out other possibilities of data deletion. 6park.com华尔街日报(The Wall Street Journal)6月28日的一篇文章含糊其辞地声称:NIH证实,应一名中国研究人员的申请删除了这些序列。这篇新闻的标题为:美国应中方要求删除新冠基因序列,病毒溯源难度加大 https://cn.wsj.com/articles/%E7%BE%8E%E5%9B%BD%E5%BA%94%E4%B8%AD%E6%96%B9%E8%A6%81%E6%B1%82%E5%88%A0%E9%99%A4%E6%96%B0%E5%86%A0%E5%9F%BA%E5%9B%A0%E5%BA%8F%E5%88%97%EF%BC%8C%E7%97%85%E6%AF%92%E6%BA%AF%E6%BA%90%E9%9A%BE%E5%BA%A6%E5%8A%A0%E5%A4%A7-11624509310 6park.comA June 28 article in The Wall Street Journal vaguely and unclearly claimed: NIH confirmed that these sequences were deleted under the request of a Chinese researcher. The headline of this news article is: The United States deletes the Sars-Cov-2 gene sequence under the request of China, making it more difficult to trace the virus to the source 6park.com令人不解的是,对如此重要的事情,华尔街日报的文章竟不说明,NIH何时证实了相关信息,证实相关信息的是NIH的哪一位工作人员,其title是什么,证实信息的有关声明发表于何处。无法判定华尔街日报的做法是有意,抑或仅仅是大意疏忽。 6park.comWhat is puzzling is that, to such an important matter, the article of WSJ does not explain when NIH confirmed the relevant information, which NIH staff member confirmed the relevant information, what its title is, and where the relevant statement confirming the information was published. It is impossible to determine whether the Wall Street Journal’s actions were intentional or merely negligent. 6park.com尽管至今仍疑点重重,但我不想过多纠缠数据到底是怎么删除的,这不是第一位的问题。以下,我将假定相关数据确实被删除了,而且确实是武汉大学项目组人员请求删除的。 6park.comAlthough there are still many doubts, I don't want to always dwell on how the data is deleted. This is not the first problem. In the following, I will assume that the relevant data has indeed been deleted, and the deletion or withdrawal is indeed requested by the Wuhan University project team. 6park.comBloom提到了一个叫Aisu Fu的人,论文说,武汉大学PRJNA612766项目的病毒样本是Aisu Fu和武汉大学人民医院搜集的。论文没有提供此人的更多信息,事实上,Aisu Fu在论文中只出现了一次。 6park.comBloom mentioned a person named Aisu Fu. The paper stated that the virus samples of Wuhan University's PRJNA612766 project were collected by Aisu Fu and Wuhan University People's Hospital. The paper did not provide more information about this person. In fact, Aisu Fu only appeared once in the paper. 6park.comAisu Fu是谁呢? 6park.comWho is Aisu Fu? 6park.comAisi Fu,中文名付爱思,是武汉臻熙医学检验实验室有限公司的总负责人,他与武汉大学药学院刘天罡教授,武汉大学人民医院李艳教授、余锂镭教授是NTS技术的共同研发者,这些信息可由以下新闻获得:武汉大学新闻网- 重磅!武汉大学联合团队开发纳米孔靶向测序 大幅提升新冠病毒阳性检出率 https://news.whu.edu.cn/info/1002/57753.htm 6park.comAisi Fu, whose Chinese name is 付爱思, is the general manager of Wuhan Zhenxi Medical Laboratory Co., Ltd. He and Professor Liu Tiangang from the School of Pharmacy of Wuhan University, Professor Li Yan and Professor Yu Lilei from the People’s Hospital of Wuhan University are Co-developers of NTS technology. This information can be obtained from the following news: Wuhan University News Network-Heavy! Wuhan University joint team develops nanopore targeted sequencing to greatly increase the positive detection rate of new coronavirus 6park.com在前面提到过的武汉大学medRxiv预印本论文中,付爱思是第二作者,不过,在那篇论文中,他的署名不是Aisu Fu,而是Aisi Fu。 https://www.medrxiv.org/content/10.1101/2020.03.04.20029538v1.full 6park.comIn the aforementioned medRxiv preprint paper of Wuhan University, Fu Aisi is the second author. However, in that paper, his signature is not Aisu Fu, but Aisi Fu. 6park.com假定Bloom恢复的SRA数据确实是武汉大学请求删除的,那么,我认为,情况应该是:NTS(纳米孔靶向测序检测)项目研发完成后,武汉大学项目组人员向NIH-SRA发出申请,撤回、删除了他们提交到SRA的数据。这些数据本来就是项目研发、测试过程中产生的非正式数据。 6park.comAssuming that the SRA data recovered by Bloom was indeed deleted under the request of Wuhan University, then I think the situation should be: After the development of the NTS (Nanopore Targeted Sequencing Detection) project is completed, the Wuhan University project team sent an application to NIH-SRA and withdrew, deleted the data they submitted to SRA. These data are originally informal data generated in the process of project development and testing. 6park.com这一撤回、删除非正式的研发、测试数据的做法有什么不正常吗?隐藏着什么不良动机吗?我看不出来。 6park.comIs there anything unusual about this withdrawal and deletion of informal R&D and test data? Are there any bad motives hidden? I can't tell. 6park.com学术论文应当专注于学术本身,基于事实,有一说一,有二说二,力求客观中立,避免被利用为政治工具,更不主动充当政治工具。妄加揣测,轻率贸然地陷人以罪,是有违科学精神,不道德,不负责任的行为。Bloom是怎么做的呢?在论文的Discussion部分,Bloom对中国科学家提出了如下指控:显然,对样本进行完全测序比偷偷删除部分序列更能提供科学信息。。。这些序列似乎很可能被删除以掩盖它们的存在。原文为:and it clearly would have been more scientifically informative to fully sequence the samples rather than surreptitiously delete the partial sequences。。。It therefore seems likely the sequences were deleted to obscure their existence. 6park.comAcademic papers should focus on the academic itself, based on facts, one says one, the other says two, strive to be objective and neutral, avoid being used as a political tool, let alone actively serving as a political tool. It is against the spirit of science, immorality, and irresponsible behavior to speculate and rashly accuse crimes. How does Bloom do it? In the Discussion section of the paper, Bloom made the following accusations against Chinese scientists: and it clearly would have been more scientifically informative to fully sequence the samples rather than surreptitiously delete the partial sequences。。。It therefore seems likely the sequences were deleted to obscure their existence. 6park.comBloom的这些话辞,是强词夺理、自相矛盾、逻辑错乱、无耻下作的造谣中伤。 6park.comBloom's words are rumors and slander of strong words, self-contradictions, logical confusion, shamelessness and despicableness. 6park.com首先,Bloom恢复的基因序列片断并非来自专门测序过程,它们是NTS(纳米孔靶向测序检测)技术研发、测试过程中产生的非正式数据,NTS主要用于医学检测、临床诊断,其输出的基因序列不是为严肃基因研究准备的。 6park.comFirst of all, the gene sequence fragments recovered by Bloom do not come from a special sequencing process. They are informal data generated during the development and testing of NTS (Nanopore Targeted Sequencing Detection) technology. NTS is mainly used for medical testing and clinical diagnosis. The genetic sequence is not prepared for serious genetic research. 6park.com第二,武汉大学在NTS研究中没有进行完全测序,不是他们故意不进行完全测序,而是因为NTS不需要进行全基因组测序,对基因组的某些重要片断进行测序就很充分了。事实上,NTS的序列检测范围,已经大大超过传统qPCR的序列比对范围,“相当于撒下了十几张大网”,同时捕捉病毒样本中的可疑基因片断。 这是NTS阳性检出率大大提高的根本原因。 6park.comSecond, Wuhan University did not perform complete sequencing in the NTS research, not because they deliberately did not perform complete sequencing, but because NTS does not need to perform whole-genome sequencing. It is sufficient to sequence some important fragments of the genome. In fact, the scope of NTS sequence detection has greatly exceeded the scope of traditional qPCR sequence comparison, "equivalent to a dozen large nets," while capturing suspicious gene fragments in virus samples. This is the fundamental reason why the positive detection rate of NTS has greatly increased. 6park.com 6park.com第三,Bloom的溯源研究论文基于一个最基本的假设:新冠病毒是自然演化产生的。如果这一假设不成立,Bloom的论文就崩溃了。Bloom一方面以武汉大学的数据作为自己自然演化理论的基础数据,基本依据,一方面又指控武汉大学的科学家偷偷摸摸删除这些可支持其自然演化理论的数据。这是一种非常错乱的逻辑。武汉大学的科学家为什么要删除“新冠自然演化”的证据?删除这些证据对中国科学家,对中国政府有什么好处?删除“新冠自然演化”的证据,掩盖“新冠自然演化”的“真相”对中国科学家,对中国政府有什么好处? 6park.comThird, Bloom's traceability research paper is based on one most basic assumption: Sars-Cov-2 is a product of natural evolution. If this assumption is not true, Bloom's paper will collapse. On the one hand, Bloom uses the data of Wuhan University as the basic data for its natural evolution theory, and on the other hand, it accuses scientists of Wuhan University of secretly deleting these data that can support its natural evolution theory. This is a very confusing logic. Why did scientists at Wuhan University delete the evidence of "natural evolution of Sars-Cov-2"? What are the benefits to Chinese scientists and the Chinese government by deleting these evidences? Deleting the evidence of the "natural evolution of Sars_Cov-2" and covering up the "truth" of the "natural evolution of Sars-Cov-2" will benefit Chinese scientists and the Chinese government? 6park.com第四,2017年12月19日,Trump政府解除了奥巴马政府3年前颁布的“功能增益研究”(Gain-of-Function, G-o-F)禁令,允许美国科学家重新申请联邦经费,开展功能增益研究,在实验室中研发、制造更具致病能力或更具传播能力的病毒或其它病原体。奥巴马禁令颁布于2014年10月22日。美国的功能增益研究与新冠病毒的出现没有关系吗?奥巴马功能增益研究禁令解除两年后,新冠病毒就出现了,这只是一种巧合吗?从2017年12月解除功能增益研究禁令,到2019年11月前后出现新冠病毒,在这近两年的时间里,美国的病毒学家们没有功能增益-改造出任何一种可怕的病毒吗? 对于Trump政府解禁功能增益研究,打开了潘多拉灾难之盒,对于美国科学家可以在联邦经费支持下,合法地在实验室功能增益-改造病毒,Bloom不置一词,装作没有这回事。事实上,对解除奥巴马禁令,重启功能增益研究,美国科学界讳莫如深,全体失声;Bllom不仅不做深刻的自我反思、自我检讨,反而扮出清白无辜的模样对中国科学家造谣中伤,栽赃嫁祸,其言行非常无耻下作。 6park.comFourth, on December 19, 2017, the Trump administration lifted the Gain-of-Function research ban promulgated by the Obama administration three years ago, allowing American scientists to reapply for federal funding to carry out G-o-F research, developing and manufacturing viruses or other Pathogens that are more pathogenic or more transmissible in the laboratory. The Obama ban was issued on October 22, 2014. Is there no relationship between the Gain-of-Function research in the United States and the emergence of Sars-Cov-2? Two years after Obama’s G-o-F ban was lifted, Sars-Cov-2 appeared. Is this just a coincidence? From the lifting of the G-o-F ban in December 2017 to the emergence of Sars-Cov-2 around November 2019, in the past two years, American virologists have not G-o-F-transformed any terrible virus? Regarding the Trump government’s lifting of the G-o-F ban, opening Pandora's disaster box, regarding American scientists, with federal funding, they can legally perform virus G-o-F and transforming, in laboratories, Bloom does not say a word, pretending that there is no such thing. In fact, the American scientific community is very secretive about lifting the Obama ban and restarting G-o-F research; instead of deep self-reflection and self-examination, Bloom instead pretends to be innocent, spreads rumors, slander, and frames Chinese scientists. His words and deeds are very shameless and despicableness. 6park.com下面讨论Bloom所恢复数据的精确性、可靠性问题。 6park.comThe following discussion is about the accuracy and reliability of the data recovered by Bloom. 6park.com由研发目的、用途及功能特点可判断,NTS检测技术是传统qPCR检测的替代技术,它的首要目的是病毒检测,而非病毒测序。即,确定样本中是否存在目标病毒是它的首要功能,测序并输出基因序列片断只是它的辅助功能。NTS技术与专门的测序明显不同,它的测序精度难以与专门测序相提并论,它的测序结果无法直接提交到国际基因数据库作为权威数据供严谨的基因研究使用。将NTS输出的基因序列片断等同于专门测序得出的权威基因组序列,并将之用于对精确度、可靠度要求极高的新冠溯源研究,我认为是不恰当的。即使这些数据确有溯源研究的价值,如线索价值,在使用时也应当非常审慎、小心,避免被数据中的错误误导。 6park.comJudging by the purpose, use, and functional characteristics, NTS detection technology is an alternative to traditional qPCR detection. Its primary purpose is virus detection, not virus sequencing. That is, determining whether the target virus is present in the sample is its primary function, and sequencing and outputting gene sequence fragments is only its auxiliary function. NTS technology is obviously different from specialized sequencing. Its sequencing accuracy is difficult to compare with that of specialized sequencing. Its sequencing results cannot be directly submitted to the international gene database as authoritative data for rigorous genetic research. I think it is inappropriate to equate the gene sequence fragments output by NTS with the authoritative genome sequence obtained by special sequencing and use it for Sars-Cov-2 traceability research that requires extremely high accuracy and reliability. Even if these data do have research value on origination of Sars-Cov-2, such as clue value, they should be used with great care and caution to avoid being misled by errors within the data. 6park.com相比qPCR,NTS技术的阳性检出率虽大大提高,达到了约75%~94%,但仍远远称不上高度精准,可以想见,它的测序功能的精准程度也是有限的;同时,NTS是疫情发生后短期内开发出来的,数据上传的2月15日,项目刚刚启动了约一个月,NTS技术尚在研发阶段,其输出的基因序列更可能存在误差、偏差甚至错误。 6park.comCompared with qPCR, the positive detection rate of NTS technology is greatly improved, reaching about 75% to 94%, but it is still far from being highly accurate. It is conceivable that the accuracy of its sequencing function is also limited; at the same time, NTS was developed within a short period of time after the outbreak of the epidemic. The data was uploaded on February 15th, at that time, and the project was just launched for about a month, NTS technology is still in the development stage, its output gene sequence is more likely to contain errors, deviations or even errors. 6park.comNTS测序功能精确度欠佳。这不是一个推测,而是一个事实。Bloom恢复、重建的基因序列片断就是这一事实的确凿证明。 6park.comThe accuracy of the NTS sequencing function is relatively poor. This is not a speculation, but a fact. The gene sequence fragments recovered and reconstructed by Bloom are conclusive proof of this fact. 6park.comBloom在论文中说:I aligned the recovered deep sequencing data to the SARS-CoV-2 genome using minimap2。。。 即:我使用minimap2比对了恢复的深度测序数据与新冠病毒基因组的一致性。minimap2是一种基因组序列比对工具。 6park.comBloom said in the paper:I aligned the recovered deep sequencing data to the SARS-CoV-2 genome using minimap2。。。 minimap2 is a genome sequence alignment tool 6park.com稍作间隔后,Bloom提供了如下表格: After a short interval, Bloom provides the following table: NTS多区段比对、检测与qPCR有限位置比对对照图 comparison chart between NTS multi-segment comparison, detection and qPCR limited position comparison
6park.com表格第一列的新冠病毒sample一共有14个,其中13个对应Bloom从SRA恢复、重建的基因序列片断,另外一个来自某个2月住院的患者。 6park.comThere are a total of 14 Sars-Cov-2 samples in the first column of the table, 13 of which correspond to the gene sequence fragments recovered and reconstructed by Bloom from SRA, and the other is from a patient who was hospitalized in February. 6park.com表格中的第二列应该是14个sample与proCov2的基因序列一致性比对结果。proCov2是天普大学(Temple University)科学家Sudhir Kumar提出的一个虚拟的新冠病毒的祖病毒。proCov2与最早发现的新冠病毒样本之一WuHan-hu-1只相差三个核苷酸,将WuHan-hu-1进行以下三个单核苷酸的更改:C8782T、C18060T和 T28144C,就得到了proCov2。 6park.comThe second column in the table should be the result of the genetic sequence consistency comparison between the 14 samples and proCov2. proCov2 is a virtual progenitor virus of Sars-Cov-2 proposed by Temple University scientist Sudhir Kumar. proCov2 is only three nucleotides apart from WuHan-hu-1, one of the first samples of Sars-Cov-2, make the following three single nucleotides modification to WuHan-hu-1: C8782T, C18060T, and T28144C, then you get ProCov2. 6park.com注1:C8782T代表:将基因序列中8782位点的胞嘧啶C对应的核苷酸(碱基对)改为胸腺嘧啶T对应的核苷酸。 6park.comNote 1: C8782T stands for: Change the nucleotide (base pair) corresponding to cytosine C at position 8782 in the gene sequence to the nucleotide corresponding to thymine T. 6park.com注2:WuHan-hu-1的基因序列是上海复旦大学张永振团队2020年1月5日上传的,是第一个上传到国际生物信息数据库的新冠病毒全基因组序列。WuHan-hu-1的病毒样本由武汉市中心医院采集提供,采集时间是2019年12月30日或26日,采集自一名41岁的陈姓新冠早期患者,该患者是华南海鲜市场的一名个体经营者。 6park.comNote 2: The gene sequence of WuHan-hu-1 was uploaded by Zhang Yongzhen's team at Fudan University in Shanghai on January 5, 2020. It is the first new coronavirus complete genome sequence uploaded to the International Bioinformatics Database. The virus sample of WuHan-hu-1 was collected and provided by Wuhan Central Hospital. The collection time was December 30 or 26, 2019. It was collected from a 41-year-old Chen surnamed early-stage patient who was Self-employed person of the South China Seafood Market. 6park.comproCov2与Wuhan-hu-1只相差3个核苷酸,而二者基因组序列(核苷酸序列)长度均为29903(含近3万个核苷酸)。易知,二者基因组序列的差异度约为0.01%(万分之一),即一致性约为99.99%。所以,Table-1中各病毒sample与proCov2的基因序列一致性比对结果,可视为这些sample与Wuhan-hu-1的一致性比对结果。 6park.comThe difference between proCov2 and Wuhan-hu-1 is only 3 nucleotides, and the length of their genome sequence (nucleotide sequence) is 29903 (containing nearly 30,000 nucleotides). It is easy to know that the difference between the two genome sequences is about 0.01% (one in 10,000), that is, the identity is about 99.99%. Therefore, the genetic sequence consistency comparison result of each virus sample and proCov2 in Table-1 can be regarded as the consistency comparison result of these samples and Wuhan-hu-1. 6park.com要注意的是,由于Bloom恢复的基因序列,也就是武汉大学NTS技术输出的基因序列不是全基因组序列,而只是全基因组序列的片断,因些,Table-1显示的不是病毒间全基因组序列的一致性,而是对应的基因组片断的一致性。Table-1标题栏已标明,比对的序列区间是21570~29550,这一区间含近8000个核苷酸。 6park.comIt should be noted that the gene sequence restored by Bloom, that is, the gene sequence output by the NTS technology of Wuhan University is not the whole genome sequence, but only fragments of the whole genome sequence. Therefore, Table-1 shows not the consistency of the whole genome sequence between viruses. but the consistency of corresponding genome fragments. The title bar of Table-1 has indicated that the sequence interval for comparison is 21570~29550, which contains nearly 8000 nucleotides. 6park.com观察各个病毒sample与Wuhan-hu-1或proCov2的一致性,可知:Table-1所列14个病毒sample中,8个与Wuhan-hu-1的一致性差别超过1%,6个超过2%,4个超过3%,3个超过4%。 6park.comObserving the consistency of each virus sample with Wuhan-hu-1 or proCov2, it can be seen that among the 14 virus samples listed in Table-1, 8 are more than 1% different from Wuhan-hu-1, 6 are more than 2%, and 4 are more than 3%, 3 more than 4%. 6park.com由于比对的基因片断含近8000个核苷酸,因此,如果序列一致性差别超过1%,那就意味着该病毒Sampe与Wuhan-hu-1至少有80个核苷酸不同; 如果一致性差别超过2%,就至少有160个核苷酸不同; 如果一致性差别超过3%,就至少有240个核苷酸不同; 如果一致性差别超过4%,就至少有320个核苷酸不同。 6park.comSince the aligned gene fragments contain nearly 8000 nucleotides, if the sequence identity differs by more than 1%, it means that at least 80 nucleotides are different between the virus sampe and Wuhan-hu-1; If it differs by more than 2%, at least 160 nucleotides are different; If it differs by more than 3%, at least 240 nucleotides are different; If it differs by more than 4%, at least 320 nucleotides are different. 6park.com比对区间外还有2万多个核苷酸位点,如果考虑这些位点,那么这些病毒sample与Wuhan-hu-1的核苷酸差异可能会更多。 6park.comThere are more than 20,000 nucleotide sites outside the comparison interval. If these sites are considered, the nucleotide differences between these virus samples and Wuhan-hu-1 may be more. 6park.com新冠病毒的变异速度是:一个病毒平均一年产生约25个核苷酸突变。产生上述规模的突变,正常情况下需要几年、十几年的时间。在疫情早期的2月15日前,武汉大学人民医院的13位患者,其体内的新冠病毒同时发生了如此显著的突变,这可能吗? 6park.comThe mutation rate of Sars-Cov-2 is: a virus produces about 25 nucleotide mutations a year on average. Under normal circumstances, it will take several to ten years to produce mutations scale above-mentioned. Before February 15 in the early stage of the epidemic, Sars-Cov-2 viruses from the bodies of 13 patients in the People's Hospital of Wuhan University had such a significant mutation at the same time, Is this possible? 6park.com对比一下迄今为止,新冠病毒的实际变异情况: 英国Alpha变种B.1.1.7的核苷酸变异位点约为28~32个; 南非Beta变种B.1.351的变异位点约为23个; 巴西Gamma变种P.1的变异位点约为17个; 印度Delta变种B.1.617.2的变异位点约为13~17个。 6park.comCompare the actual mutation of the new coronavirus so far: There are about 28-32 nucleotide variation sites in the British Alpha variant B.1.1.7; There are about 23 mutation sites in South Africa Beta variety B.1.351; There are about 17 mutation sites in the Brazilian Gamma variety P.1; There are about 13-17 mutation sites in the Indian Delta variety B.1.617.2. 6park.com可见,疫情发展至今,四个最重要的新冠病毒变种,其变异位点数都小于40个核苷酸;相比之下,Bloom恢复的数据有8个呈现出了超过80个核苷酸位点的不同,有3个呈现出了超过320个核苷酸位点的不同。而且,这么大幅度的核苷酸差异发生在去年2月15日前发现的病毒样本中。 6park.comIt can be seen that the four most important Sars-Cov-2 variants so far have less than 40 nucleotide sites; in contrast, 8 of the data recovered by Bloom show more than 80 nucleotide sites. Three of them showed a difference of more than 320 nucleotide positions. Moreover, such a large nucleotide difference occurred in virus samples discovered before February 15 last year. 6park.com这些核苷酸位点的差异都是突变造成的吗?如果是的话,如此超常的突变,Bloom怎么会视而不见呢?这个困惑昨天未能解决。今天,我返回Bloom的论文,下载了几个Bloom恢复、重建的基因序列,将它们的基因序列与Wuhan-Hu-1的基因序列加以比对(使用NCBI Blast工具),而后发现:原来,绝大多数核苷酸差异对应着核苷酸缺失,与变异无关。Bloom重建序列中存在着大量的字母N,每一个字母N都代表着其所在位点的核苷酸缺失。 6park.comAre these differences in nucleotide positions all caused by mutations? If so, how could Bloom turn a blind eye to such an extraordinary mutation? This confusion could not be resolved yesterday. Today, I returned to Bloom’s paper, downloaded several Bloom’s restored and reconstructed gene sequences, and compared their gene sequences with the Wuhan-Hu-1 gene sequence (using NCBI Blast tool), and then found that: it turns out, Overwhelming majority nucleotide differences correspond to nucleotide deletions and have nothing to do with variation. There are a large number of letters N in the Bloom reconstruction sequence, and each letter N represents a nucleotide deletion at its location. 6park.comBloom重建序列中的大量核苷酸缺失,要么是Bloom重建序列时产生的,要么是武汉大学NTS测序错误造成的。 6park.comA large number of nucleotide deletions in Bloom's reconstructed sequence are either caused by Bloom's reconstruction of the sequence, or due to sequencing errors of NTS technology of Wuhan University. 6park.com对所采用的基础数据的准确性、可靠性问题,以及它们对研究过程、论文结论可能造成的影响,Bloom在论文中没有进行讨论和评估。 6park.comBloom did not discuss and evaluate the accuracy and reliability of the basic data used, and their possible impact on the research process and the conclusion of the paper. 6park.com(正文完) 6park.com相关文章: 6park.com病毒学家Bloom溯源论文的基础数据问题 https://club.6parkbbs.com/bolun/index.php?app=forum&act=threadview&tid=15952734 6park.com虚假伪劣、贼喊捉贼,打假美国之音溯源新发现 https://club.6parkbbs.com/bolun/index.php?app=forum&act=threadview&tid=15950371 6park.com中国科学家是否删除了新冠早期基因组数据? https://club.6parkbbs.com/bolun/index.php?app=forum&act=threadview&tid=15949751 6park.com附录:Bloom重建的基因序列片断示例。 Appendix: Examples of gene sequence fragments reconstructed by Bloom
https://club.6parkbbs.com/chan2/index.php?app=forum&act=threadview&tid=13521676 6park.com该示例对应Table-1中的C9,即第四个病毒Sample,序列中的N代表核苷酸缺失的位点。该示例是C9基因序列片断的一部分,未展示全部近8000个核苷酸(碱基对)。 6park.comThis example corresponds to C9 in Table-1 of the main post, which is the fourth virus Sample. The N in the sequence represents the site of nucleotide deletion. This example is part of a fragment of the C9 gene sequence and does not show all of the nearly 8000 nucleotides (base pairs). 6park.comBloom论文提供了13个重建新冠基因序列片断的下载地址: https://github.com/jbloom/SARS-CoV-2_PRJNA612766/raw/main/results/consensus/consensus_seqs.csv 6park.comThe Blooms‘ paper provides the download address of 13 reconstructed sequence fragments of Sars-Cov-2.: 6park.com(全文完) 6park.com贴主:苦难与荣耀于2021_07_05 3:46:19编辑 6park.com贴主:苦难与荣耀于2021_07_05 5:35:10编辑评分完成:已经给 苦难与荣耀 加上 100 银元!
6park.com贴主:苦难与荣耀于2021_07_05 23:37:10编辑
贴主:苦难与荣耀于2021_07_15 22:49:13编辑